Introduction
In this article, we introduce mass spectrometry testing as a new and promising technology for blood-based MRD in multiple myeloma. We will discuss the two different mass spectrometry methods currently available and highlight their benefits and limitations.
Multiple myeloma is a plasma cell disorder caused by the buildup of myeloma cells in the bone marrow. These cancerous cells produce high levels of abnormal monoclonal immunoglobulin proteins, called M-spike.
In 2006, the International Myeloma Working Group (IMWG) published a uniform response criteria for multiple myeloma. Patients who reach a complete response (CR) do not have a detectable M-spike. This is determined by standard blood or urine tests which include serum protein electrophoresis (SPEP) and immunofixation (IFE). New treatment regimens have increased response rates close to 100%. Complete response rates are now over 70%. The goal of minimal residual disease (MRD) testing is to uncover signs of disease that might be missed by traditional tests. This helps doctors better assess how well the patient is responding to treatment.
According to the IMWG (Kumar, 2016), MRD negativity means there are no myeloma cells detected in a sample of 100,000 cells. Studies show that MRD negativity is linked to better progression-free survival (PFS) and overall survival (OS), with patients achieving MRD negativity at the 10-6 threshold showing the greatest improvement.1 However, even MRD negative patients can eventually relapse. This highlights the need for more sensitive and reliable tools to better measure residual disease at a lower level.
Bone Marrow MRD Tests
Gold standard MRD tests, such as next generation sequencing (NGS) or next generation flow cytometry (NGF), track MRD by measuring the number of abnormal plasma cells, also known as myeloma cells, in the bone marrow. However, obtaining bone marrow samples is a painful and invasive procedure. This limits how often these tests can be used.
Due to the patchy nature of myeloma, obtaining high quality bone marrow samples remains particularly challenging. Clinicians will use imaging tests, like PET scanning or MRI, to improve the success of obtaining a viable sample. Even so, there is still a risk of false negative test results due to the effects of hemodilution or poor sampling technique.
Some limitations include the need to identify a baseline clone for NGS-based MRD tests. However, a suitable baseline clone may not be available in 3% to 20% of samples.2 In next generation flow cytometry (NGF), the sensitivity of the test can vary between the different labs that perform the test. Additionally, NGF tests can only be performed on freshly pulled samples, which must be processed within a relatively short 48 hour window.3
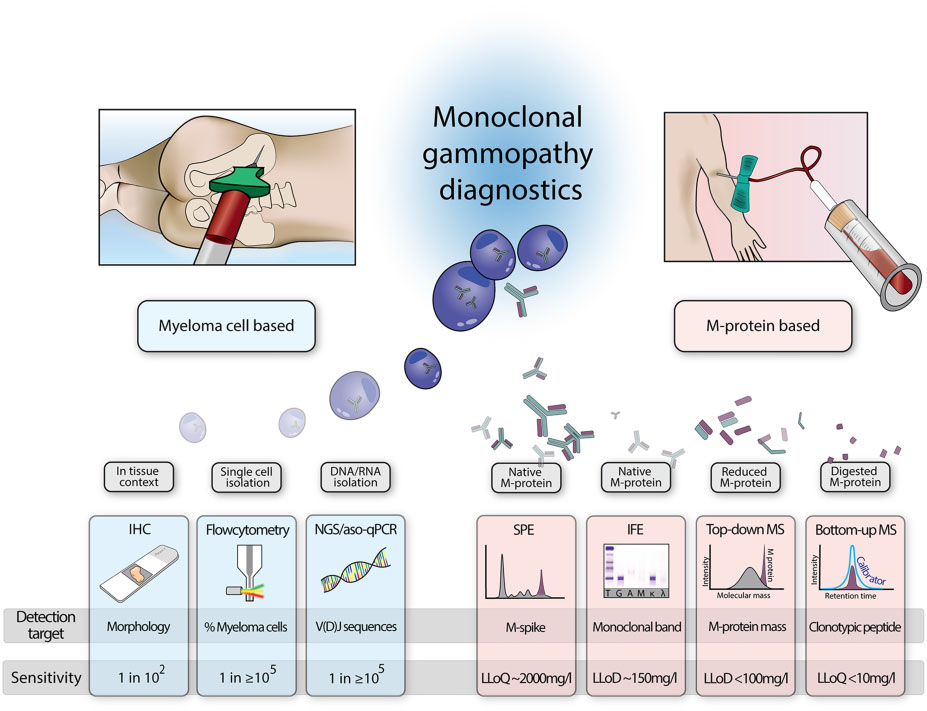
Fig 1. Overview of different techniques to monitor monoclonal gammopathy disease activity. IHC, immunohistochemistry;
LLoD, lower limit of detection; LLoQ, lower limit of quantification. Zajec et al. Clin. Chem. 2020
To address these gaps, clinicians are looking to new highly sensitive blood tests for multiple myeloma that are able to detect and track M-spike to concentrations 1000x lower than the limit of detection of SPEP.4 The ability to test directly from peripheral blood without the need for invasive and painful bone marrow aspirates overcomes many of the limitations of bone marrow-based assays and opens new opportunities for more frequent and routine MRD monitoring in the clinic.
Mass Spectrometry Blood Tests in Multiple Myeloma
Mass spectrometry (MS) is an analytical technique that identifies molecules, such as a patient’s M-spike, based on its mass-to-charge ratio. Every protein has a unique sequence of amino acids which gives the protein its own unique mass and charge. MS tests are able to identify and quantify proteins based on this unique ratio of mass to charge.
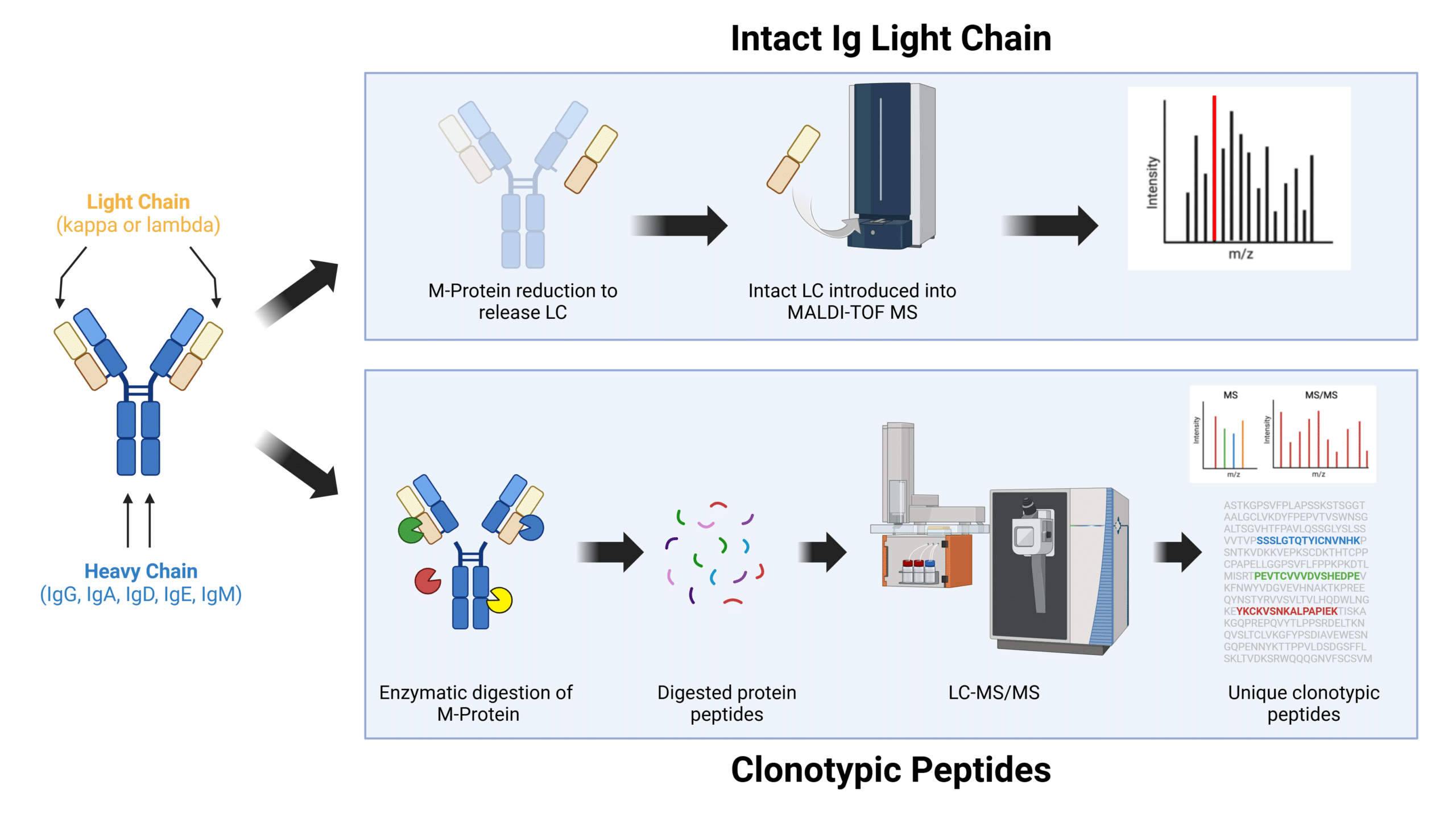
The two main approaches to MRD testing by MS in multiple myeloma are 1) intact immunoglobulin (Ig) light chain and 2) clonotypic peptides.
Fig. 2. Different MS approaches to M-protein monitoring in MM. LC-MS/MS, liquid chromatography tandem mass spectrometry. MALDI-TOF, matrix assisted laser desorption ionization-time of flight mass spectrometry. LC, light chain.
Intact Immunoglobulin Light Chain (MALDI-TOF)
In the intact Ig light chain approach, the intact M-spike is first reduced to break down the disulfide bonds to release the light chain (LC) from the heavy chain (HC). The intact LC protein is then directly analyzed by a MALDI-TOF mass spectrometer. Little preparation is required and testing can be done quickly and without the need for a baseline.
Despite being more sensitive than an SPEP (0.1g/dL) or IFE (0.05g/dL), the sensitivity of the intact Ig mass approach can only reliably monitor M-spike down to a concentration of 0.01 g/dL.5 Below this level, the M-spike cannot be reliably identified from the noise of the polyclonal background (ie. the signal of the background mixture of other antibodies circulating in the body). Another drawback is that some antibody therapeutics can share a similar molecular profile to the patient’s M-spike light chain and can cause interference.
Clonotypic Peptides Mass Spectrometry (LC-MS/MS)
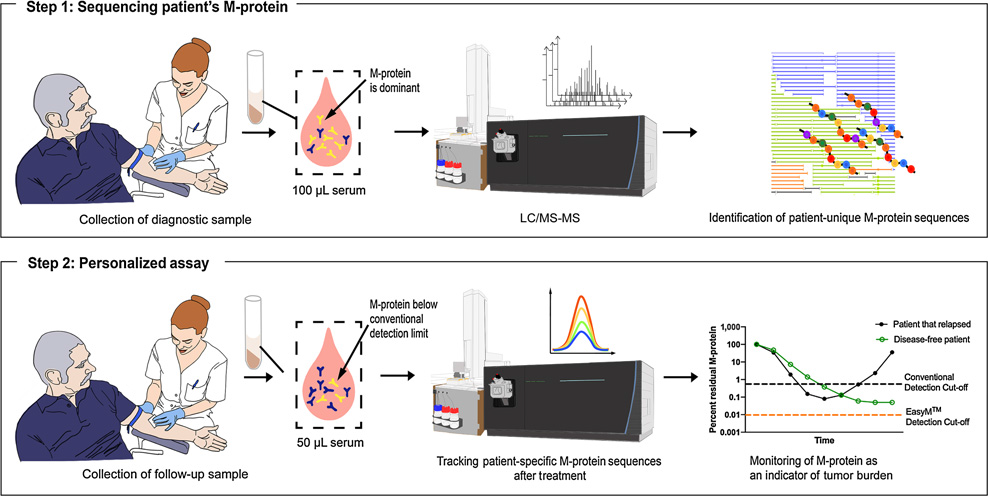
The clonotypic peptides mass spectrometry approach overcomes many of these drawbacks. In this method, the M-spike is first digested by enzymes into smaller protein fragments, called peptides, before being separated by size and charge through a liquid chromatography column. The sorted peptides are then analyzed by the mass spectrometer to determine its amino acid sequence, some of which are unique to the patient. These unique peptides are selected, screened and subsequently used in follow up tests to identify and monitor M-spike concentrations.
The greatest advantages of the clonotypic peptide approach over the intact Ig mass approach are that it is 100% patient specific and significantly more sensitive, being able to detect M-spike as low as 0.00001 g/dL. Put differently, this is over 1000x more sensitive than an SPEP.
In a study comparing EasyM, a clonotypic peptides mass spectrometry test, to bone marrow-based EuroFlow NGF MRD assay, EasyM was able to detect and quantify M-spike in 53% of samples that were negative by NGF.6 Another advantage of the clonotypic peptides approach is that it is 100% patient specific and not susceptible to interference from therapeutic antibodies, making it well suited for the ongoing monitoring of MRD.7 The main limitations are that it is a time consuming assay to perform and requires a suitable baseline serum sample early in treatment.
Limitations
A limitation shared by all MS-based tests is that they are not useful in non-secretory MM patients who do not produce an M-spike protein. Another limitation is that M-spike proteins are naturally recycled in the body over time. This means that during the treatment phase, even in the absence of active myeloma cells, an M-spike signal can still linger in the blood. After 1 to 6 months, depending on the isotype, the residual M-spike proteins will eventually be cleared from the body.
One study in newly diagnosed myeloma patients showed that a >99% reduction in M-spike levels from baseline at the Day 100 post-transplant time point was found to be prognostic.7 Such thresholds could be useful in assessing treatment response at early time points with mass spectrometry blood tests where the M-spike has not yet completely disappeared. More work is needed to further validate these thresholds at early time points.
References
- Munshi NC, Avet-Loiseau H, Anderson KC, et al. A large meta-analysis establishes the role of MRD negativity in long-term survival outcomes in patients with multiple myeloma. Blood Adv. 2020;4(23):5988–5999. doi: 10.1182/bloodadvances.2020002827.
- Ferla V, Antonini E, Perini T, et al. Minimal residual disease detection by next-generation sequencing in multiple myeloma: promise and challenges for response-adapted therapy. Front Oncol. 2022;12:932852. doi: 10.3389/fonc.2022.932852
- Charalampous C, Kourelis T. Minimal residual disease assessment in multiple myeloma patients: minimal disease with maximal implications. Front Oncol. 2022;11:801851. doi: 10.3389/fonc.2021.801851
- Liyasova, M. et al. A Personalized Mass Spectrometry-Based Assay to Monitor M-spike in Patients with Multiple Myeloma (EasyM). Clin Cancer Res. 2021. 15;27(18):5028-5037. doi: 10.1158/1078-0432.CCR-21-0649.
- Murray, DL. et al. Mass spectrometry for the evaluation of monoclonal proteins in multiple myeloma and related disorders: an International Myeloma Working Group Mass Spectrometry Committee Report. Blood Cancer J. 2021. 11(2):24. doi: 10.1038/s41408-021-00408-4.
- Zhao J, Khong T, Gorniak M, et al. Comparison of EasyM, a clonotypic mass spectrometry assay, and EuroFlow minimal residual disease assessment in multiple myeloma. Haematologica 2024 Nov 7. doi: 10.3324/haematol.2024.285933
- Slade, M. et al. Measurable Residual Disease Status By Clonotypic Mass Spectrometry with Easym Assay Predicts Outcomes Following Autologous Hematopoietic Cell Transplant in Multiple Myeloma. Blood 2023. 142 (Supplement 1): 3354. doi: https://doi.org/10.1182/blood-2023-172440
- Kubicki, T. et al. Mass spectrometry-based assessment of M protein in peripheral blood during maintenance therapy in multiple myeloma. Blood. 2024 Aug 29;144(9):955-963 doi: 10.1182/blood.2024024041.